Page 11 - DMN1Q22
P. 11
Introduction
In mid-November of 2021, a novel variant of the SARS-COV-2 virus caused a sharp increase in the number of people testing positive for COVID-19 in Southern Africa and Botwswana. Due to the concerns related to the number and type of mutations in this variant’s genome, the World Health Organization and the Centers for Disease Control and Prevention (CDC) classified this novel variant as a variant of concern (VOC). It was named the ‘Omicron’ variant in the WHO’s Greek-alphabet system for VOCs. The first case of the Omicron variant was identified in the United States on December 1, 2021. Within a few weeks, it had become the dominant variant across the globe- the fast-
est variant replacement in the history of the SARS-COV-2. By January 1, 2022, the United States witnessed over half a million COVID-19 cases each day. In mid-January of 2022, one of the more transmissible sublineages of the Omicron variant (BA.2) came to the notice of public health authorities in Europe, espe- cially in Denmark and the United Kingdom. BA.2 was highlight- ed in the lay media as a ‘stealth’ variant, a somewhat confusing and incorrect terminology. Intended for healthcare professionals, this essay summarizes the information on Omicron’s mutational profile and pathophysiology, and covers the early information around B.2.
Based on their biological and epidemiological properties, public health organizations classify significant variants as variants being monitored, variants of interest, variants of concern, and variants of high consequence.
Each SARS-CoV-2 virus has 24-40 haphazardly arranged spikes (or S-protein) that tend to bind to the ACE-2 receptors on human cells. The S-protein contains a receptor-binding subunit (called S1) and a fusion subunit (called S2). First, the S protein binds to the host receptor through the receptor-binding domain (RBD) in the S1 subunit. After that, Furin and Furin-like en- zymes act on polybasic cleavage sites at the S1-S2 junction and cleave the two subunits from each other. Once the S1-S2 site has been cleaved, a second cleavage site on S2 becomes accessible
to other proteases. Once this site is cleaved, the S2 can fuse with the host membrane and undergo conformational changes that bring the viral and host membranes together.
The S-protein is a crucial pathological protein in COVID-19, re- sponsible for viral binding to the ACE-2 receptor and serving as the main target of neutralizing antibodies elicited during natural infection and post-vaccination. Any mutations in the genetic regions encoding RBD, S1-S2 cleavage sites, and the base of the S-protein can significantly alter the function of the S-protein and consequently modify the transmissibility and pathogenicity of the novel variant.
Mutations in the Omicron Variant
Since its discovery in South Africa, scientists and public health organizations have been concerned by the constellation of mutations in the Omicron variant- many previously associated with hyper-transmission and immune evasion. It has the highest number of mutations in its genome among the VOCs, more than 60. Of all Omicron mutations, 50 are nonsynonymous, eight are synonymous, and two are non-coding. A nonsynony- mous modifications can change the amino acid sequence in the encoded protein- causing changing in the biological behavior of the protein.
Omicron’s mutations are spread across its entire genome. Specif- ically, the S-protein region has over 30 mutations. Many occur in the critical RBD region on residues 319-531 and the cleavage sites on S1 and S2. For example, the G339D, K417N, N440K, and E484A mutations confer some resistance to monoclonal therapies and convalescent sera - and are thus associated with immune escape capabilities. In addition, the N501Y mutation, nicknamed “Nelly,” has been seen in other VOCs.
Along with T478K and Q498R mutations, the N501Y mutation changes the electrostatic environment inside the RBD. It creates additional contact points for RBD and ACE2. These changes increase the RBD-ACE2 affinity. Discovered at the same time as Nelly, the D614G mutation, nicknamed “Doug,” had spread across the world in late 2020 and has been seen in other VOCs. Doug makes the hinge region on the S-protein more flexible, improving viral transmission. Finally, the H655Y, N679K, and P681H mutations are located near the Furin cleavage site. All
of them improve the efficiency of protein cleavage, aiding in the entry of the virus into the host cell. In the long term, this improved entry efficiency enables increased transmission rates.
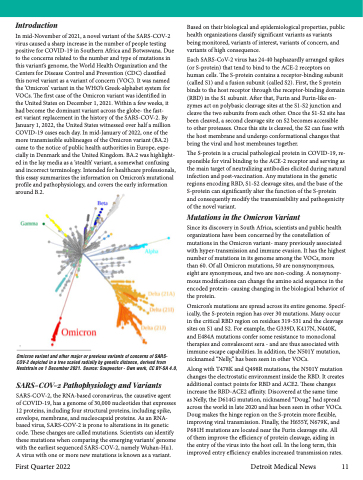
Omicron variant and other major or previous variants of concerns of SARS- COV-2 depicted in a tree scaled radially by genetic distance, derived from Nextstrain on 1 December 2021. Source: Soupvector - Own work, CC BY-SA 4.0,
SARS-COV-2 Pathophysiology and Variants
SARS-COV-2, the RNA-based coronavirus, the causative agent of COVID-19, has a genome of 30,000 nucleotides that expresses 12 proteins, including four structural proteins, including spike, envelope, membrane, and nucleocapsid proteins. As an RNA- based virus, SARS-COV-2 is prone to alterations in its genetic code. These changes are called mutations. Scientists can identify these mutations when comparing the emerging variants’ genome with the earliest sequenced SARS-COV-2, namely Wuhan-Hu1. A virus with one or more new mutations is known as a variant.
First Quarter 2022
Detroit Medical News 11